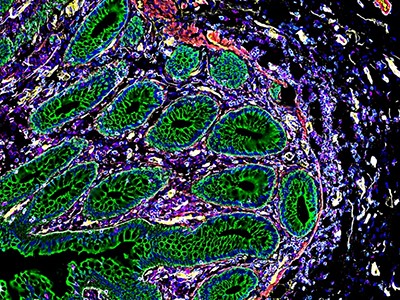
A large collection of papers maps the biology and locations of a range of cancer cells, including colon cancer (pictured).Credit: Steve Gschmeissner/Science Photo Library
Detailed maps that pinpoint the positions of cells in tumours and probe the tumours’ biology are offering insights into the development of several cancers — including in the breast, colon and pancreas — and could provide leads for potential treatments.
In a tranche of 12 papers published in Nature journals on 30 October, researchers of the Human Tumor Atlas Network (HTAN) analysed hundreds of thousands of cells from human and animal tissues. Some of the studies describe 3D maps of the cells — known as cell atlases — in tumours, whereas others create ‘molecular clocks’ that trace the cellular changes that lead to cancer.
“Applying these novel tools to cancer allow us to look at them with a different lens,” says Ken Lau, a computational cell biologist at Vanderbilt University Medical Center in Nashville, Tennessee, and co-author of a study that records the timing of cellular events in the development of colorectal cancer1. “We can actually see things that we couldn’t see before.”
Mapping tumours
In some of the studies, researchers created atlases that allowed them to study tumours at single-cell resolution and investigate how cancer begins. One team analysed the organization of cells in 131 samples of six types of cancer, including those of the breast, colon, pancreas and kidney2. The scientists found that distinct regions in the same tumour could respond differently to drugs. Understanding how various cancer-cell clusters respond to treatments could help researchers to design more effective ones.
Cell ‘atlases’ offer unprecedented view of placenta, intestines and kidneys
Other studies used 3D mapping to study samples of colon polyps — abnormal growths in the lining of the gut that can turn cancerous. They identified molecular changes in the polyps’ cells, including the loss of DNA connections and alterations in the activity of genes3, as well as changes in the immune response, cell growth and hormone metabolism4 that occur early on and could cause the polyp cells to turn cancerous.
Therapies that target these changes could make cancer treatments and early health interventions more effective, says Ömer Yilmaz, a stem-cell biologist at the Massachusetts Institute of Technology in Cambridge. “The best treatment for cancer is prevention. And if we can understand how different cell populations respond to the environment and to the diet, how that impacts tumour initiation and how different clones contribute to that process, it could lead to better prevention or detection methods.”
Immunity insights
Other atlases offer clues about why some cancer types are more challenging to treat than others. “Tumours are not just composed of cancer cells,” says Daniel Abravanel, a physician–scientist at the Dana-Farber Cancer Institute in Boston, Massachusetts, and co-author of a study on breast cancer5. For instance, immunotherapies, which don’t target cancer cells directly but aim to help the immune system eliminate them, are less effective against breast cancer than other types, he adds.
To investigate why, Abravanel and his colleagues created a 3D tumour atlas using dozens of samples from 60 people with aggressive forms of breast cancer. They studied how immune cells are distributed and found that some types of immune cell were more common than others in certain tumours, especially in people who had received immunotherapy.
Why are so many young people getting cancer? What the data say
For three people, biopsies taken from the same tumour 70–220 days apart showed differences in the levels of immune cells known as T cells and macrophages. In two cases, the numbers of these cells had decreased over time, whereas in the third they had increased.
This “really shows how dynamic the immune microenvironment is, and it may explain why attempts to characterize tumours and predict responses to immune-checkpoint therapies from one biopsy at a single time point has had inconsistent results”, says Brian Lehmann, a breast-cancer researcher who specializes in genomics at the Vanderbilt-Ingram Cancer Center in Nashville, Tennessee.
In another study, researchers found that some aggressive subtypes of breast cancer had more immune cells than others, and seemed to become ‘worn out’ over time6. These cells expressed a protein called CTLA4, which limits their ability to respond to tumours. Therapies targeting CTLA4 have shown promising results in treating melanoma and lung cancer. “This opens up additional avenues for using that therapy in a subset of breast cancers,” says Lehmann.
CRISPR clock
Other experiments reveal insights into how cells turn cancerous in the first place. In the colorectal-cancer study, Lau and his colleagues engineered a ‘molecular clock’ to track how normal cells start to turn rogue and begin proliferating uncontrollably in the gut1. They used single-cell analysis and a gene-editing CRISPR tool to generate mutations in each cell’s DNA. These mutations acted as time stamps, recording the timeline of changes in and divisions of each cell.
The race to map the human body one cell at a time
Lau and his team applied this approach to 418 human colon polyps and found that up to 30% of the polyps originated from several cell types, rather than from a single cell. In 60% of the polyps, one group of cells started to ‘overtake’ others as the polyp grew — developing into a tumour. Two similar studies in mice7,8, including an analysis of 260,922 single cells from 112 samples of intestinal tissues, also found that a mix of cells collectively initiate colorectal tumours.
These findings overturn previous thinking that colon cancers arise from single rogue cells in the gut’s lining, and could open up opportunities for early diagnosis and intervention.
“To evaluate the risk of [pre-cancerous growths], what people use is the size. The larger the tumour, the more risk it has,” says Lau. But the molecular-clock and other analyses reveal that “there may be other biomarkers in terms of looking at genetics and evolution”.