Mapping eQTLs and sQTLs at high resolution
MAGE offers a valuable resource for uncovering the genetic factors that drive variation in gene expression and splicing, including genetic variation that is largely private to historically underrepresented populations. By intersecting published genotype data from the same set of samples24, we mapped cis-eQTLs and cis-sQTLs within 1 Mb of the transcription start site (TSS) of each gene. We define eGenes and sGenes as genes with an eQTL or sQTL, respectively, and eVariants and sVariants as the individual genetic variants defining an eQTL or sQTL signal, respectively. We note that although we performed QTL mapping for genes on the autosomes and the X chromosome, we focus on results from the autosomes here owing to several methodological details that are specific to the X chromosome (Supplementary Methods). Across 19,539 autosomal genes that passed expression-level filtering thresholds (Supplementary Methods), we discovered 15,022 eGenes and 1,968,788 unique eVariants (3,538,147 significant eVariant–eGene pairs; 5% false discovery rate (FDR)). Additionally, across 11,912 autosomal genes that passed splicing-filtering thresholds, we discovered 7,727 sGenes and 1,383,540 unique sVariants (2,416,177 significant sVariant–sGene pairs; 5% FDR).
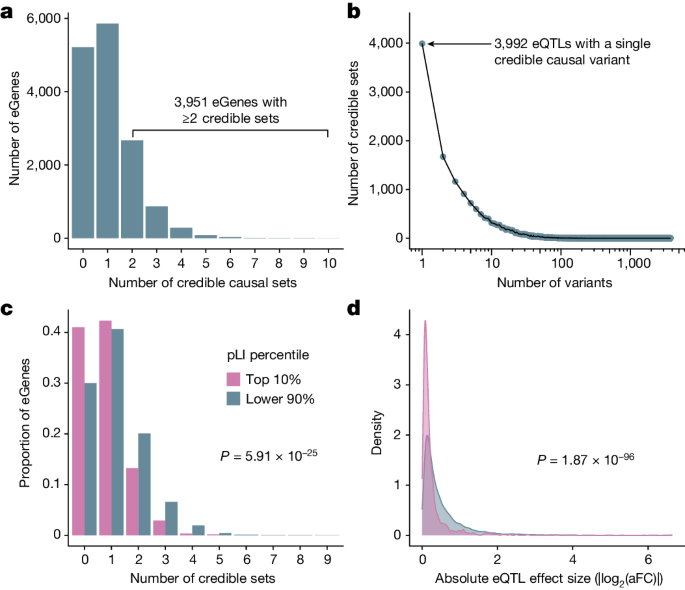
The inclusion of genetically diverse samples in association studies reduces the extent of LD and improves mapping resolution8,10 (Supplementary Fig. 11). With this advantage in mind, we used SuSiE25 to perform fine mapping for all eGenes and the introns of all sGenes to identify causal variants that drive each QTL signal. For each gene and intron, SuSiE identifies one or more credible sets, representing independent causal eQTL and sQTL signals and whereby each credible set contains as few variants as possible while maintaining a high probability of containing the causal variant. To obtain a gene-level summary of the sQTL fine-mapping results, we collapsed intron-level credible sets into gene-level credible sets by iteratively merging intron-level credible sets for each sGene (Supplementary Methods). We identified at least one credible set for 9,807 (65%) eGenes and 6,604 (85%) sGenes, which we define as fine-mapped eGenes and sGenes, respectively. Consistent with previous results4,26,27, we observed widespread allelic heterogeneity across fine-mapped genes, with 3,951 (40%) of fine-mapped eGenes and 3,490 (53%) of fine-mapped sGenes exhibiting more than one distinct credible set (Fig. 3a and Extended Data Fig. 2c). We also achieved high resolution in identifying putative causal variants that drive expression changes. That is, of 15,664 eQTL credible sets, 3,992 (25%) contained a single variant (median 5 variants per credible set; mean = 15.8, s.d. = 65.7; Fig. 3b). Similarly, for sQTLs, 3,569 out of 16,451 (22%) credible sets contained a single variant (median 7 variants per credible set; mean = 23.6, s.d. = 99.1; Extended Data Fig. 2d). For downstream analyses, we selected a single representative ‘lead QTL’ from each eGene and sGene gene-level credible set.
a, Number of credible sets per eGene, demonstrating evidence of widespread allelic heterogeneity, whereby multiple causal variants independently modulate expression of the same genes. b, Fine-mapping resolution, defined as the number of variants per credible set. c, A signature of negative selection against expression-altering variation, whereby eGenes under strong evolutionary constraint (defined as the top pLI decile reflecting intolerance to loss-of-function mutations; pink) possess fewer credible sets, on average, than other genes (blue). d, A signature of negative selection against expression-altering variation, whereby eQTLs of genes under strong evolutionary constraint (top pLI decile; pink) have smaller average effect sizes (aFC) than other genes (blue).
For each lead eQTL, we calculated its effect size using an implementation of the allelic fold change (aFC)28 statistic that quantifies eQTL effect sizes conditional on all other lead eQTLs for that gene (Supplementary Methods). We observed that 2,031 (13%) lead eQTLs had a greater than twofold effect on gene expression (median |log2(aFC)| = 0.30; mean = 0.51, s.d. = 0.64; Extended Data Fig. 1). This was a slightly smaller proportion than previously reported by GTEx26, but we propose that this is partially explained by the small sample sizes in some GTEx tissues, which drives a stronger ‘winner’s curse’, whereby effects are systematically overestimated29.
Evidence of selective constraint
Previous studies of large population cohorts have identified sets of genes under strong mutational constraint, whereby negative selection has depleted loss-of-function point mutations and copy number variation30. One metric for quantifying mutational constraint on genes is the probability of intolerance to loss-of-function mutations (pLI)30. In our data, we observed that eGenes possessed significantly lower mean pLI scores (mean = 0.261, s.d. = 0.395) than non-eGenes (mean = 0.304, s.d. = 0.409; two-tailed Wilcoxon rank-sum test: W = 11,596,590, P = 3.89 × 10−7). Additionally, highly constrained eGenes (top 10% of pLI) tended to possess fewer credible sets (mean = 0.80, s.d. = 0.82) than other eGenes (mean = 1.12, s.d. = 1.04; two-tailed quasi-Poisson generalized linear model: \(\hat{\beta }\) = −0.354, P = 5.91 × 10−25; Fig. 3c). Moreover, the average effect size of lead eQTLs within highly constrained genes (mean |log2(aFC)| = 0.25; s.d. = 0.36) was smaller than that of other genes (mean |log2(aFC)| = 0.53; s.d. = 0.65; two-tailed Wilcoxon rank sum test: W = 3,789,053, P = 1.87 × 10−96; Fig. 3d). This difference was apparent regardless of whether the minor allele is associated with higher (Δmean |log2(aFC)| = −0.277; two-tailed Wilcoxon rank-sum test: W = 928,592, P = 1.39 × 10−50) or lower expression (Δmean |log2(aFC) = −0.268; two-tailed Wilcoxon rank sum test: W = 967,228, P = 2.97 × 10−47), consistent with a model of stabilizing selection whereby gene expression is maintained within an optimal range. These results indicate an association between constraint against loss-of-function protein-coding sequence variation (that is, pLI) and constraint against expression-altering variation (that is, number of credible sets and eQTL effect sizes). This association held for several other metrics of mutational constraint that include intolerance to copy number variation (that is, pHaplo and pTriplo) as well as divergence-based estimates of sequence conservation in putative promoter elements (Extended Data Fig. 3). Together, our results are consistent with previous analyses demonstrating weak, but measurable, selection against expression-altering variation31.
Functional enrichment of QTLs
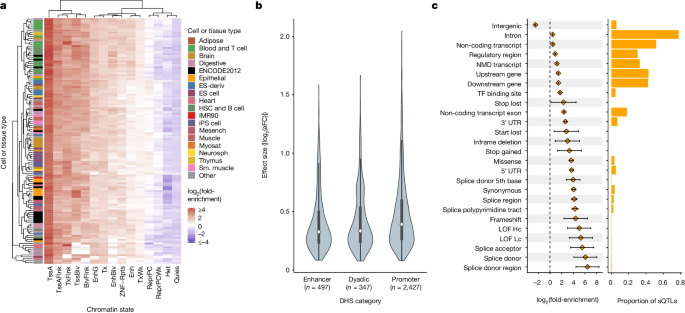
Taking advantage of the high resolution of putative causal signals, we quantified the enrichment of fine-mapped lead eQTLs in 15 predicted chromatin-state annotations across 127 reference epigenomes from the Roadmap Epigenomics chromHMM model32. Enrichment was most pronounced within promoter regions, specifically at active TSSs (TssA) and flanking regions (TssAFlnk), but modest enrichments were also apparent within enhancer regions (Enh and EnhG), especially for blood cell types (Fig. 4a and Supplementary Fig. 12B). Conversely, quiescent, repressive and heterochromatic regions were depleted of eQTLs. We further extended our analysis to primary DNase hypersensitivity site (DHS) annotations, and we observed a strong enrichment of lead eQTLs in DHSs of blood and T cell samples (Supplementary Fig. 12C).
a, A heatmap representing hierarchical clustering of the enrichment of cis-eQTLs in predicted chromatin states using the Roadmap Epigenomics 15-state chromHMM model across 127 cell and tissue samples. b, Distribution of absolute value of lead cis-eQTL effect sizes measured as log2(aFC) across putatively active chromatin states of LCLs linked to multi-tissue DHSs. Sample sizes describe the number of unique eVariants annotated as belonging to each of the DHS categories. Bars represent the first, second (median) and third quartiles of the data and whiskers are bound to 1.5× the interquartile range. c, Enrichment of lead sQTLs (n = 13,107 unique sVariants total, at least 5 per category) within functional annotation categories from Ensembl Variant Effect Predictor (left), along with the proportion of all lead sQTLs falling into each annotation category (right). Error bars denote 95% CI around the estimated sQTL enrichment in each category. Enrichment was calculated in comparison to a background set of variants matched on MAF and distance from the TSS. Annotation categories are not mutually exclusive and therefore sum to a proportion greater than 1. ES, embryonic stem; HSC, haematopoietic stem cell; iPS, induced pluripotent stem; Mesench, mesenchymal cell; Myosat, myosatellite cell; Neurosph, neurosphere; Sm., smooth; TssA, active TSS; TssAFlnk, flanking active TSS; TxFlnk, transcription at gene 5′ and 3′; Tx, strong transcription; TxWk, weak transcription; EnhG, genic enhancer; Enh, enhancer; ZNF/Rpts, ZNF genes plus repeats; Het, heterochromatin; TssBiv, bivalent/poised TSS; BivFlnk, flanking bivalent TSS/enhancer; EnhBiv, bivalent enhancer; ReprPC, repressed polycomb; ReprPCWk, weak repressed polycomb; Quies, quiescent/low; NMD, nonsense-mediated mRNA decay; LOF, loss of function; Hc, high confidence; Lc, low confidence.
Focusing on data from LCLs, we next explored the relationship between epigenomic enrichments and eQTL effect sizes (|log2(aFC)|). Promoter-associated enrichment was consistent across eQTL effect size deciles, and enrichment within poised regulatory regions such as bivalent TSS (TSSBiv) and bivalent enhancers (EnhBiv) was most apparent for eQTLs of large effect sizes (Supplementary Fig. 13A,B). By contrast, eQTLs located within chromatin states associated with transcribed regions (Tx, TxWk and TxFlnk) predominantly exhibited lower effect sizes (Supplementary Fig. 13C). These qualitative trends were replicated in other primary blood cell types (Supplementary Figs. 14–17). Using additional DHS-based annotations from Roadmap Epigenomics32, we observed larger median eQTL effect sizes in promoter regions relative to enhancers and dyadic (that is, acting as both promoter and enhancer) regions (Fig. 4b). This pattern was similarly replicated across other primary blood-related cell types (Supplementary Figs. 14–17). Using chromatin immunoprecipitation followed by sequencing data from ENCODE33, we also observed that lead eQTLs were significantly enriched within 312 (92.30%; Bonferroni-adjusted P < 0.05) transcription factor (TF) binding sites, including canonical promoter-associated TFs such as POLR2A, TAF1, JUND, ATF2 and KLF5, as well as TFs such as HDACs, EP300 and YY1, which are typically associated with enhancers (Supplementary Fig. 12A).
We also investigated the genomic context of our fine-mapped cis-sQTLs. We observed strong enrichment of lead sQTLs in several key splicing-relevant annotations, including splice donor sites (log2(fold enrichment) = 6.07, 95% confidence interval (CI) = 4.09–8.04) splice acceptor sites (log2(fold enrichment) = 5.52, 95% CI = 3.54–7.50) and nearby regions (log2(fold enrichment) = 4.15, 95% CI = 3.70–4.62) at intron–exon boundaries (Fig. 4c). Despite their magnitude of enrichment, variants in canonical splice sites and splice regions represented a minority of lead sQTLs, with a greater abundance of sQTLs falling within 5′ and 3′ untranslated regions (UTRs), as well as exons of both coding and noncoding genes. Although exhibiting weaker enrichments, these annotation categories together covered a much larger mutational target size and may encompass splicing enhancers and cryptic splice sites. By contrast, intergenic regions were strongly depleted of lead sQTLs (log2(fold enrichment) = −2.51, 95% CI = −2.58 to −2.43). Together, these findings provide support for the biological validity of the fine-mapped cis-QTLs and insight into the mechanisms by which these variants affect gene expression and splicing.
Colocalization of eQTLs and sQTLs and GWAS hits
To explore the role of expression-associated genetic variation in human complex traits, we next sought to discover shared signals between fine-mapped MAGE cis-eQTLs and cis-sQTLs and results from genome-wide association studies (GWAS). As a multi-ancestry resource, we anticipate that MAGE will facilitate the interpretation of GWAS from underrepresented populations. One such cohort is the Population Architecture using Genomics and Epidemiology (PAGE) study8, which comprises 49,839 non-European individuals, including large samples of individuals who self-reported as Hispanic/Latin American or African American, as well as smaller samples of individuals who self-reported as Asian, Native Hawaiian or Native American. We performed colocalization analysis to identify shared signals between GWAS of 25 complex traits from PAGE and cis-eQTLs and cis-sQTLs from MAGE. PAGE GWAS data include quantitative biomedical traits such as platelet count and cholesterol levels, as well as diseases such as type 2 diabetes (see Supplementary Table 1 for a full list of the traits included in this analysis).
Across these 25 traits, we identified 384 independent GWAS signals. For each independent GWAS signal, we tested for eQTL colocalization with each eGene within 500 kbp. We implemented this analysis using a combination of SuSiE25 and coloc34,35 to allow for multiple causal variants at each signal and to allow for different patterns of LD between the two datasets. We defined moderate colocalizations as those with posterior probabilities ≥ 0.5 and strong colocalizations as those with posterior probabilities ≥ 0.8.
Using this approach, we identified moderate colocalizations with MAGE cis-eQTLs for 39 independent GWAS signals across 14 traits and strong colocalizations for 25 independent GWAS signals across 13 traits (Supplementary Fig. 18). These included 6 GWAS signals across 6 traits for which the GWAS variant was rare (minor allele frequency (MAF) < 0.05) or unobserved in the European continental group in the 1KGP. Among these, one notable result involved colocalization (Pcoloc = 0.998) between a platelet count GWAS hit (sentinel variant rs73517714) and an eQTL hit of the tropomyosin gene TPM4, whereby the lead eQTL variant (rs143558304) falls within the 3′ UTR. Previous work has implicated rare missense variants in TPM4 with platelet abnormalities and excessive bleeding36, findings that provide support for a role of this gene in platelet function. The MAGE lead eQTL and the GWAS sentinel variant were in strong LD (R2 = 0.874 in MAGE) and were rare (MAF < 0.05) in the European continental group of the 1KGP but more common in the African continental group.
We repeated this colocalization analysis for MAGE sQTLs. Across the same set of 384 GWAS signals, we identified moderate colocalizations with MAGE cis-sQTLs for 30 independent GWAS signals across 12 traits and strong colocalizations for 24 independent GWAS signals across 10 traits (Supplementary Fig. 18). These included three GWAS signals across two traits for which the GWAS variant was rare or unobserved in the European continental group in the 1KGP. Together these results highlight the utility of paired globally diverse gene expression and WGS datasets like MAGE and 1KGP, respectively, in interpreting complex trait GWAS of non-European cohorts.