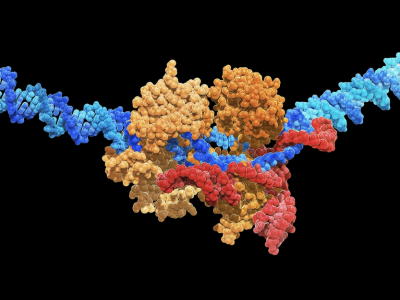
From roundworms to humans, signalling through the Notch protein-receptor family drives embryonic development, cellular differentiation and tissue homeostasis. It’s also absolutely essential for the transformation of immature human immune cells into T cells — cellular warriors that target viruses and tumours.
Yet the activation of Notch signalling has been hard to mimic in the laboratory, making it close to impossible to exploit in the clinic and hampering efforts to generate T-cell therapies. But that could be changing.
In July, attendees at a conference in Lewiston, Maine, dedicated to Notch signalling were treated to not one, but two new tools to activate the pathway. Unbeknown to each other, two research groups had been working in parallel to develop what they unveiled at the conference.
“It was a turning point,” says Juan Carlos Zúñiga-Pflücker, a developmental immunologist at the University of Toronto in Canada, whose lab has pioneered many Notch activation techniques. For him, the announcements represented a “sort of a re-emergence” of this work in the Notch field.
Trouble with 2D
Most receptors undergo some kind of conformational change after binding to their ligands, leading to a chain of events that alters the cell’s behaviour. But ligand binding alone isn’t enough to activate Notch.
Once the receptor binds to its ligand (which is tethered to another cell), the ligand-expressing cell reels in the ligand-receptor complex. Like tugging at a loose thread in a sweater, the resulting pulling force unravels part of the Notch receptor, exposing it to enzymes that release the receptor’s intracellular domain and allow it to travel to the nucleus to influence gene expression.
Stealthy stem cells to treat disease
“The real technological challenge is coming up with a drug that can mimic that pulling effect,” says Vincent Luca, a protein engineer at the Moffitt Cancer Center in Tampa, Florida.
Current strategies require the co-culture of cells expressing the Notch receptor with cells that express the ligand, or fixing the ligand onto plates or beads. But these ‘2D’ methods can’t be used in vivo, says Luca.
They’re also a bottleneck for large-scale T-cell production, says George Daley, a haematologist and stem-cell biologist at Harvard Medical School in Boston, Massachusetts, because only the cells at that ligand-presenting interface can be activated. For years, Daley’s lab has struggled with the limitations of 2D methods while trying to coax pluripotent stem cells into T cells.
In 2021, Daley was approached by Rubul Mout, then a postdoctoral researcher at the University of Washington in Seattle in the lab of David Baker, who won a share of the 2024 Nobel Prize in Chemistry for his work in computer-aided protein design. Mout wanted to apply those design techniques to Daley’s stem-cell-based systems.
Daley was “tremendously excited” at the idea, and before Mout even officially joined his lab, they discussed engineering a soluble Notch agonist — one that could present a Notch ligand to cells in suspension rather than on a flat surface.
Turbocharged CAR-T cells melt tumours in mice — using a trick from cancer cells
Using the Rosetta protein-design tool developed in Baker’s lab, Daley, Mout and their colleagues designed potential agonists by attaching multiple copies of the ligand to a protein scaffold. They experimented with designs with as few as two and as many as 120 copies of the ligand, as well as different geometries.
One arrangement worked particularly well: three copies of the ligand radiating out from the scaffold, like spokes. When added to Notch-expressing cells in suspension, this multivalent ligand linked the cells together in a microscopic pas de deux, with each developing T cell gently pulling on (and activating) the other. Compared with 2D activation, Notch activation using the soluble agonist in a bioreactor created five times more T cells per microgram of agonist1, suggesting that this approach might be useful for large-scale T-cell manufacturing.
Boosting Notch
Luca says that his group, by contrast, was searching for a “targeted, focused way of activating Notch”, with an eye to therapeutics. For example, boosting Notch could rejuvenate exhausted T cells trying to destroy a tumour — but this must be done without activating the tumour cells, which also express Notch.
Luca’s lab generated SNAGs, synthetic Notch agonists (see ‘Mimicking notch’), by fusing a gene for a Notch agonist to one that encodes an antibody fragment targeting cancer cells. The result encodes a bispecific protein that forms what Luca calls a “little molecular bridge” between a Notch-expressing cell and the cancer-antigen-expressing cell. That binding event mechanically activates the receptor in a manner akin to natural Notch signalling2.
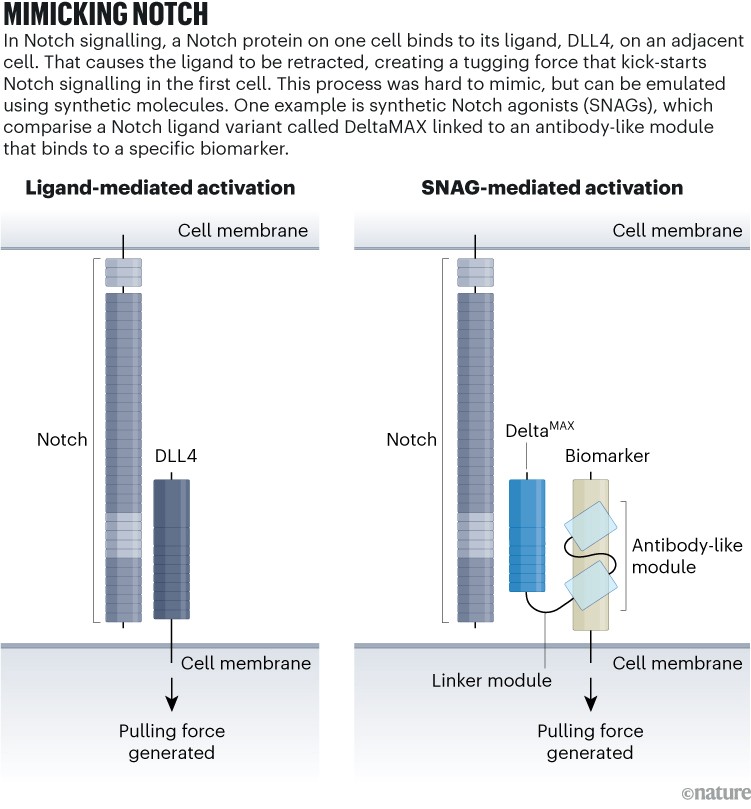
Source: Adapted from ref. 2
A very different soluble Notch agonist was unveiled in January. Biophysicist Björn Högberg and his team at the Karolinska Institute in Stockholm study the minuscule, tactile interactions between proteins during cell signalling, which Högberg likens to “a Braille system” that the cells use to talk to one another.
Working with Luca, the team created a hybrid DNA-protein agonist by attaching multiple copies of the Notch ligand along a piece of DNA that had been folded, origami-style, into a rod. To their surprise, the agonist activated Notch without generating any measurable pulling force3.
The researchers suggest that their origami agonist might be triggering an alternative mechanism of activation, in which long-term ligand binding is sufficient to cause receptor unravelling, albeit at a much slower pace. But they’re still trying to understand how this might work.
Highly mutated cancers respond better to immune therapy
Reactions from the community about the possibility of a pulling-independent Notch activation mechanism have been mixed, Högberg says: “I think some of them were really intrigued and really, really wanted to know more, and some of them were like, ‘I don’t think this is anything.’”
For his part, Zúñiga-Pflücker is sceptical that the origami ligand works without any sort of pulling force. But “the work is really nicely done”, he says, adding that “all three papers are quite exciting in different ways”.
Even just a few years ago, says Mout, de novo protein design wasn’t advanced enough to address meaningful biology. But the field is rapidly evolving, he says, and real problems are being solved. “Everything is coming together now,” he says.